abundance sorting
See also
UCLUST algorithm
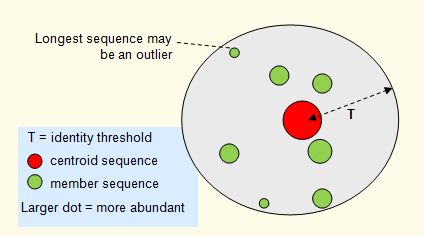
Centroids and sort order
sortbysize command
Global trimming
Abundance sorting is a pre-processing step for clustering when
a more abundant sequence is likely to be the best centroid (representative
sequence) for a cluster. This is often the case, for example, in 16S OTU clustering of next-generation sequencing reads. In this case, more abundant sequences are likely to be accurate biological sequences while rare or singleton reads are more likely to contain sequencing errors or be due to PCR artifacts such as chimeras.
See assigning reads to OTUs for more
discussion.
It is strongly recommended that amplicon read sequences are globally trimmed before clustering and abundance sorting.
In older versions of QIIME, CD-HIT was the default OTU clustering method.
CD-HIT sorts by decreasing length, which is a bad choice for OTUs because the
longest read is often an outlier and tends to have more errors, especially
with 454 reads.