Machine learning in OTU analysis
 See also
See also
Random forests
k-fold cross-validation
OTU
importance
otutab_forest_train command
otutab_forest_classify command
otutab_forest_kfold command
Machine learning refers to automated methods for creating
classifiers. A classifier is an algorithm which assigns categories to
observations, where an observation is described by numerical values for a
set of features. For
example, categories could be "man" and "woman", and the data could be a
photograph of a face. Here, features are pixels, and values represent
colors.
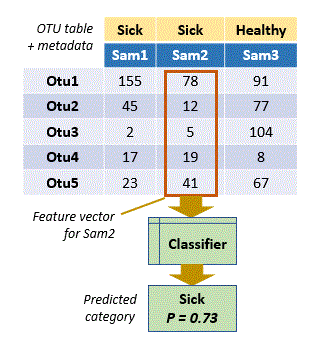
In OTU analysis, observations are samples and categories
are metadata such as healthy / sick, day /
night, and so on. The data describing the observation is the set of OTU
counts or frequencies in a sample, i.e. a column in the OTU table. The column can be viewed as a
vector, which is machine learning terminology would be called a feature
vector (OTUs are features).
With unsupervised learning, the algorithm attempts to
infer patterns in the data without reference to pre-assigned labels or
categories. This type of algorithm is rarely (never?) used in OTU analysis.
If you know of an application of unsupervised learning here, please let me
know.
With supervised learning, the parameters of a classifier are trained on
observations which are labeled with their correct categories. This is called
"supervised" because the machine needs to be helped (supervised) during the
learning phase, but not during the classification phase.
A trained classifier can be used to predict categories of novel
samples. This could be used, for example, to create a diagnostic test for a
gut disorder using 16S data from stool samples. However, this approach is rarely used in practice.
If you know of a good example, please let me know.
The most common use of machine learning in
OTU analysis is to answer the question: does the composition of the
community change with the sample metadata state (healthy / sick etc.)? Put
another way, can metadata states be predicted from OTU counts or
frequencies? This question can be answered by using
k-fold cross-validation on samples with known metadata.



 See also
See also