Cross-talk example: Caporaso et al. data
See also
Cross-talk
uncross command
Samples are from a mock community and seven different environments. The top 25 OTUs generated by UPARSE are shown sorted by decreasing number of reads. Table entries shown in orange are probably due to cross-talk. This is most obvious in the mock samples as most OTUs do not match the designed community (blue RefSeqs). See below for further discussion.
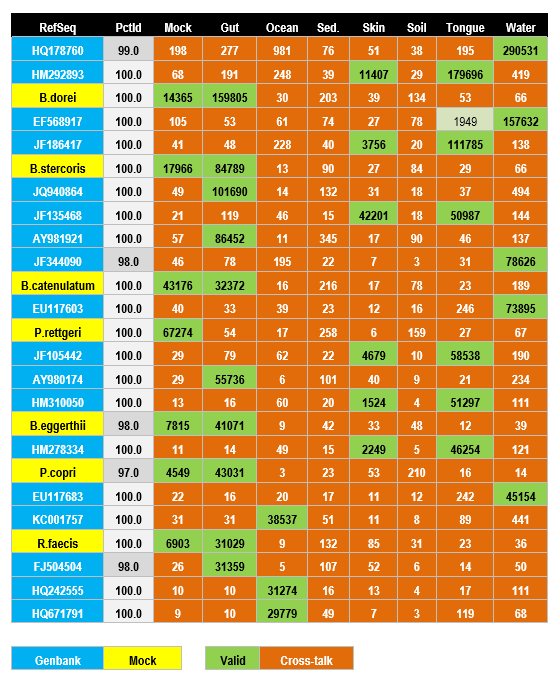
The top 25 most abundant OTUs from a multiplexed GAIIx run. Most entries in this table are probably spurious due to cross-talk and should therefore be zero. OTUs are sorted in order of decreasing total abundance. Counts are manually annotated as valid (green) or cross-talk (orange). Most cases are readily classified except Tongue in the fourth OTU (light green) which has 1949/159979=1.2% of the total reads, which could be cross-talk but is a distinctly higher fraction than other probable cross-talk counts seen in the table. Reference sequences with species names (yellow) are designed strains in the mock community, otherwise are Genbank identifiers (blue). Nine species are missing from the mock reference database, so some, but not all, of the OTUs marked with Genbank identifiers may be expected mock OTUs. PctId gives the OTU identity with the reference sequences. Two of the mock identities are <100% which is probably due to reference sequences which do not match exactly because they were obtained from different strains of the same species. Data from Caporaso et al. 2011.