Cross-validation by identity
See also
TAXII benchmark
Microbial taxonomy
sintax command
nbc_tax command
Top-hit identity distribution
distmx_split_identity command
Cross-validation by identity (CVI) is a benchmark
strategy for assessing accuracy of predictions of features or traits from
genes or gene fragments such as 16S sequences. In the
TAXXI benchmark, it was used to
measure accuracy of taxonomy prediction.
Motivation
CVI explicitly models
varying distances between query sequences and the reference database. This
is because in practice, OTUs have a range of identities with the reference.
OTU identities are sometimes low, creating a challenging problem for
prediction algorithms. For example, with taxonomy prediction, only ~20k 16S sequences
are currently known from isolate strains with reliably known
taxonomies, and OTUs often have low identities (say, 90% and lower) with
these sequences. Prediction is more difficult at lower identities, but in
most previous
benchmark tests query sequences usually have high
identities with the reference and thus fail to model a realistic scenario.
The range of identities between a given set of OTUs and a reference database
can be summarized as a top-hit identity histogram.
 Implementation
Implementation
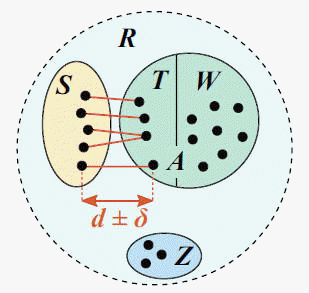
The distmx_split_identity command
implements CVI is as follows. A reference database with known traits (e.g.,
taxonomy) is split into test and training sets such that for all test
sequences, the most similar training sequence has a given identity (top hit
identity, d), e.g. d=97%
(see figure). R is the reference database, which is
divided into disjoint subsets S, T,
W and Z. S
is the test set; the training set is A =
T + W. T
is the set of top hits for sequences in S, which
are constrained to be in the range d +/- delta
where delta specifies the maximum allowed deviation from the
desired identity d. W
contains reference sequences with identity < d;
these are retained to create the largest possible training set.
Z contains sequences which cannot be assigned to S,
T or W without violating the
identity constraint.
Making a benchmark dataset
Construct
test-training pairs for several different identities. This enables assessment of accuracy at
varying distances from the reference. For example, with taxonomy, high accuracy at family
rank is expected for query sequences having 100% identity with the reference
database, but lower accuracy at 90%; these expectations can be validated by
test/training pairs at 100% and 90% identities, respectively. Query
sequences belonging to novel taxa, i.e. taxa not found in the reference, are
modeled in test/training pairs with d < 100%. For example, most pairs of 16S
rRNA sequences in a given genus have >= 95% identity. Therefore, with
d = 90%
most test sequences will belong to genera which are absent from the training
set, and with d = 95% there will be a mix of present and absent genera. Thus,
novel taxa arise naturally by construction of the test/training pairs, and
the frequency of novel and known taxa at each rank is determined by identity,
which can be measured for any OTU,
rather than taxonomy, which is not known.
References (please cite)
R.C. Edgar (2018), Accuracy of taxonomy prediction for 16S rRNA and fungal ITS sequences, PeerJ 6:e4652
• Cross-validation by identity, novel benchmark strategy enabling realistic accuracy estimates
• Genus accuracy of best methods is 50% on V4 sequences
• Recent algorithms do not improve on RDP Classifier or SINTAX
R.C. Edgar (2018), Taxonomy annotation and guide tree errors in 16S rRNA databases, PeerJ 6:e5030
• Approx. one in five SILVA and Greengenes taxonomy annotations are wrong
• SILVA and Greengenes trees have pervasive conflicts with type strain taxonomies



 Implementation
Implementation